The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein has been the main target for the development of vaccines and other therapeutics needed to treat COVID-19 illness. New research published in the journal Future Medicine found the SARS-CoV-2 spike glycoprotein has druggable cavities with the best target being compatible with more than 1 billion compounds.
One compound known as Compound 38 had the highest affinity to a SARS-CoV-2 cavity and prevents viral entry. Results suggest it meddles with the interaction between the SARS-CoV-2 receptor binding domain and the human ACE2 receptor.
Compound 38 could be a potent inhibitor for blocking SARS-CoV-2 and may help shape future COVID-19 therapy.
SARS-CoV-2 Druggable Cavities. Image Credit: https://www.futuremedicine.com/doi/10.2217/fvl-2020-0394
How they did it
The researchers performed a computational analysis of the SARS-CoV-2 glycoprotein to search for potential binding sites. The search was facilitated by a program called CavityPlus which predicted the 3D structure of the spike glycoprotein. A SARS-CoV-2 cavity was considered ‘druggable’ if it was associated with a high drug score during pharmacophore modeling.
A virtual screening of a library with a large number of natural chemical compounds was used to improve the odds of finding a candidate that could target SARS-CoV-2 cavities. Autodock Vina was used to measure compounds with the highest affinities toward the druggable cavities.
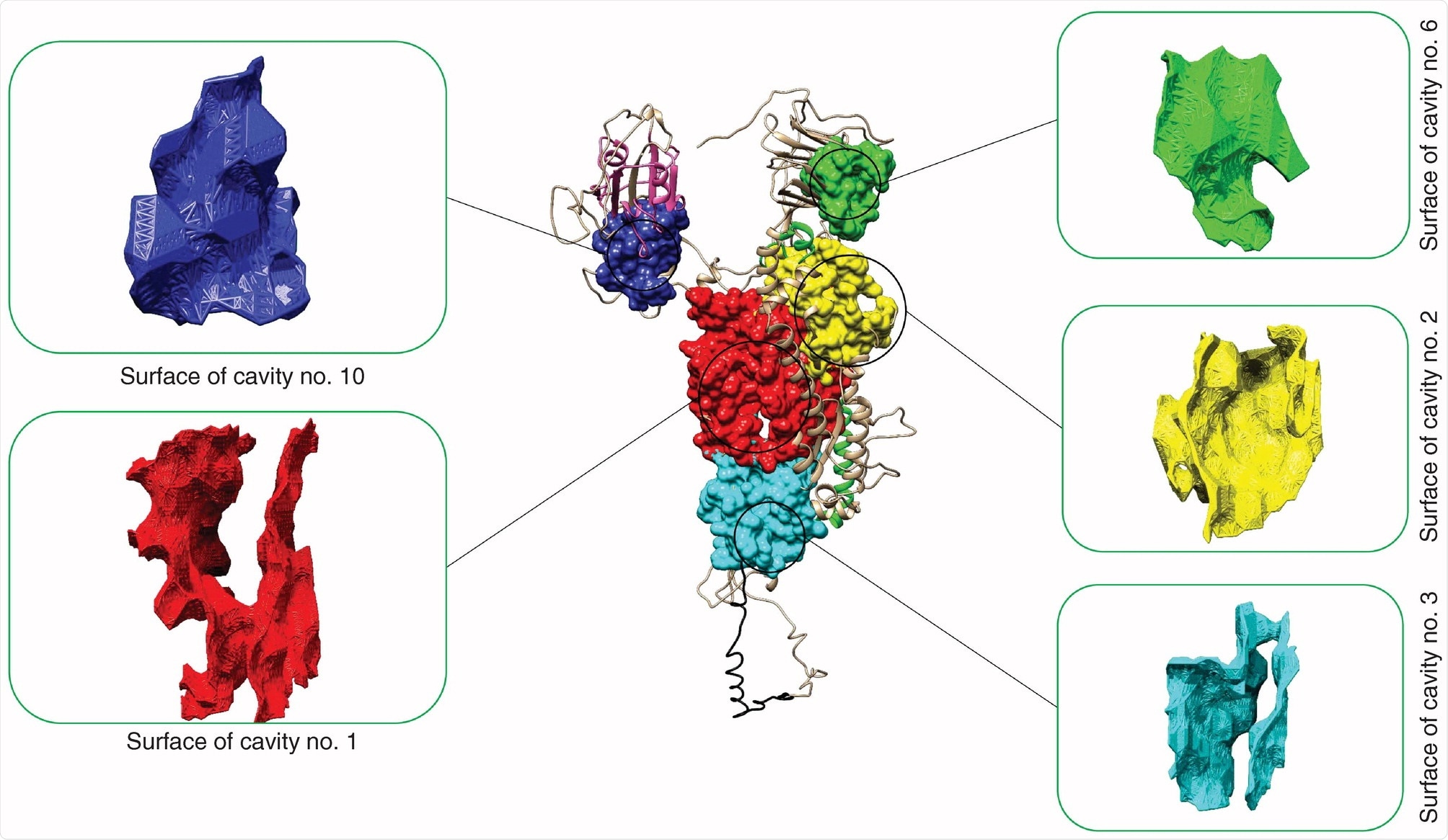
SARS-CoV-2 cavity number 10 has the greatest ‘druggable’ potential
Forty-two SARS-CoV-2 cavities were identified in the spike glycoprotein with five being potentially druggable. When searching the SARS-CoV-2 spike glycoprotein, the team found one druggable cavity next to the receptor-binding domain of the S glycoprotein. The druggable cavity number 10 had the highest drug score during pharmacophore modeling and was used for the rest of the study.
Of more than 1 billion compounds, 410 were potential matches for drug cavity number 10.
Compound 38 blocks SARS-CoV-2 infection
Of the 410 chemical compounds tested, compound 38 had the highest affinity for the drug cavity because of interactions with amino acids G381, E516, L517, L518, H519, R567, D571, and T572 in the virus glycoprotein. Amino acid residues E516, L517, and D571 had the closest contact with the cavity.
The compound 38 helped block viral entry with no significant toxicity. The finding was confirmed when observed an increase in affinity when substituting nitrogen N4 with CCN and H functional groups of the compound to the receptor.








 User Center
User Center My Training Class
My Training Class Feedback
Feedback












Comments
Something to say?
Log in or Sign up for free